Data is loading...
How to use
Search 6mers
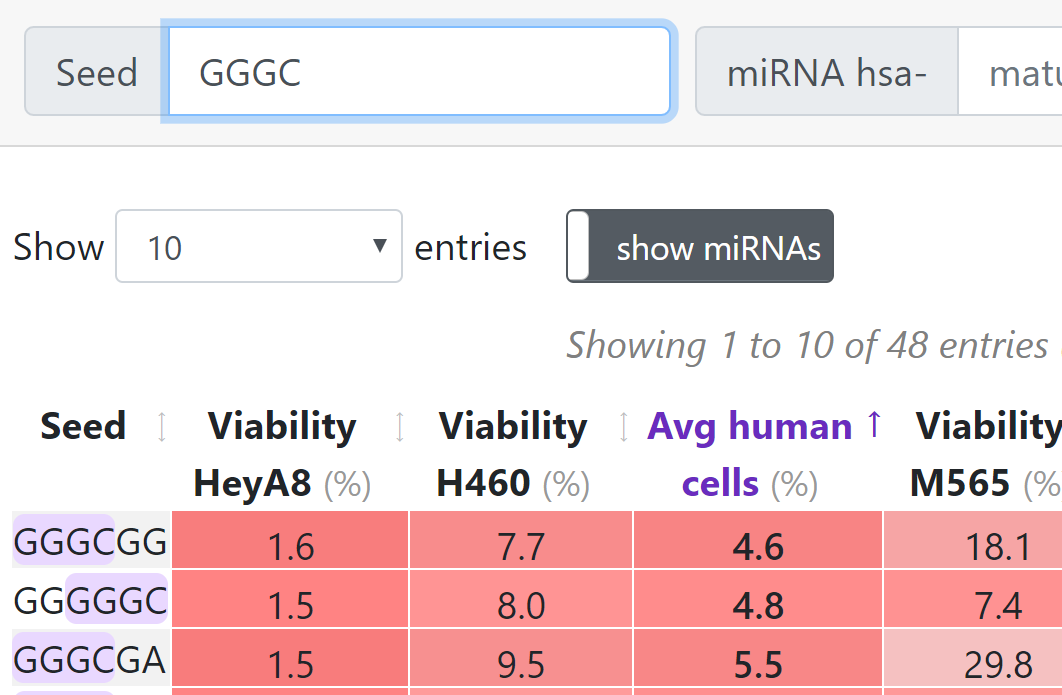
Enter one of 4096 possible 6mers into the search field or any nucleotide sequence of less than 7 nucleotides.
Use positions 2-7 of the guide/antisense strand of the RNA duplex as the 6mer.
Search miRNAs
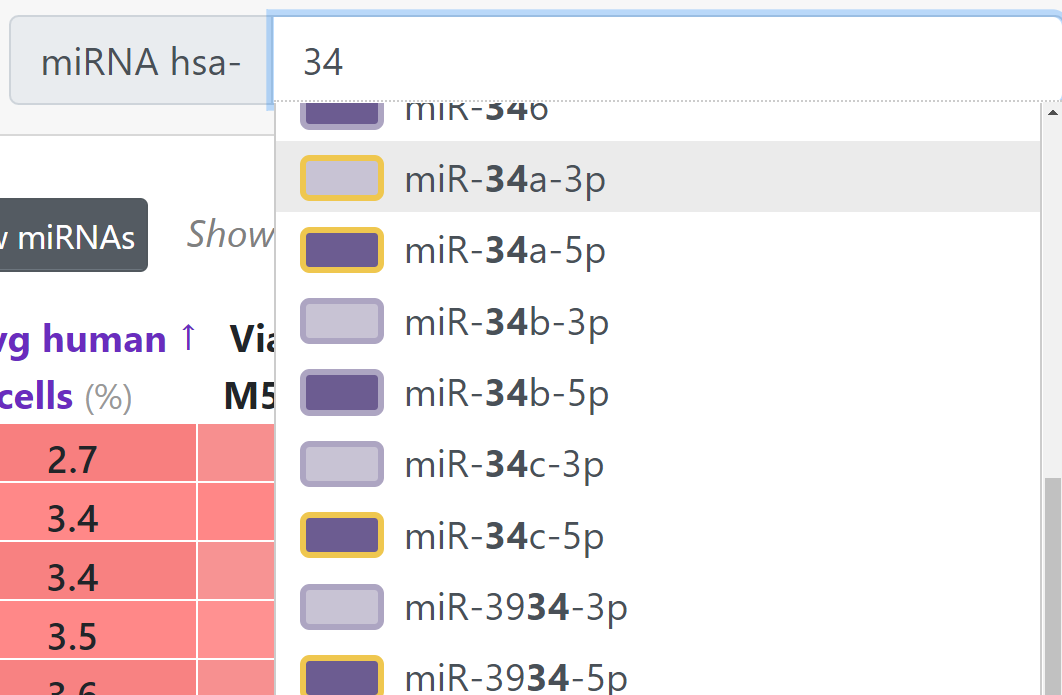
Type in the miRNA search field. It will auto-complete to show matching mature miRNAs. You can also use the miRNA accession number.
Each miRNA is color-coded. The colors are explained in the Info section below.
miRBase links
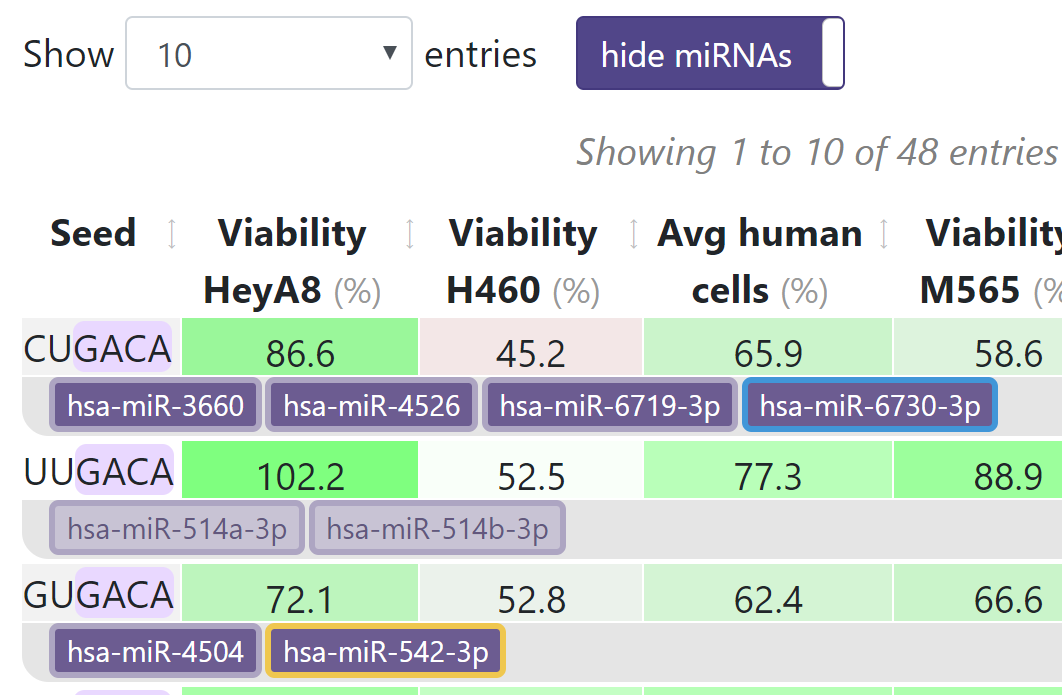
With show miRNAs enabled, you can click on a mature miRNA in the results to be taken to the mirbase.org page corresponding to its accession number.
Info
This site allows to determine the activity of any siRNA, shRNA, or miRNA to be toxic through its 6mer seed sequence in human (HeyA8 - ovarian, H460 - lung and H4 - brain) and mouse (M565 - liver, 3LL - lung and GL261 - brain) cancer cell lines.
The viability columns of the data table are colored as follows:
- miRNAs with low viability (high toxicity, red color) will likely be tumor suppressive.
- miRNAs with high viability (low toxicity, green color) can be tumor promoting.
When show miRNAs is enabled, miRNAs are color-coded as follows:
- Mature miRNAs representing the predominant arm are shown in dark purple.
- Mature miRNAs representing the lesser expressed arm (the * activity) are shown in light purple.
- miRNAs that are highly expressed and processed by Drosha (RISC bound in wild-type but not Drosha k.o. HCT116 cells) have a yellowoutline.
- All other miRNAs are either not processed by Drosha (i.e. miR-320a), miRtrons, or not significantly expressed in HCT116 cells.
- miRtrons have a blueoutline.
References
Gao, Q.Q., Putzbach, W.E., Murmann, A.E., Chen, S., Sarshad, A.A., Peter, J.M., Bartom, E.T., Hafner, M. and Peter, M.E. (2018) 6mer seed toxicity in tumor suppressive microRNAs. Nature Comm, 9:4504.
Version History
Click for detailed changelogs.
Acknowledgements
This site was developed by Johannes Peter.
The source code is available on Github under the MIT license
You are welcome to file Bug Reports or make Feature Requests